Single Cell Approach Reveals Impact of Disease-Causing Gene Mutations
Post Date: March 22, 2021 | Publish Date: April 22, 2020

Our aim is to create a general workflow that can be applied to many diseases. To achieve this, we leverage new genomics approaches and computational strategies to find out which mutations, cell-types, developmental periods, and molecular pathways are likely driving the disease.”
— Nathan Salomonis, PhD
Changes in DNA can have very different consequences—some cause disease, while others have no effect. So how can we determine which mutations are important and which aren’t?
Single-cell methods hold the key to finding answers, according to a breakthrough study in the journal Nature led by H. Leighton Grimes, PhD, of the Division of Immunobiology, and Nathan Salomonis, PhD, of the Division of Biomedical Informatics.
The study introduces a new workflow to tackle the challenge of linking gene mutations
with disease-causing processes. Researchers and clinicians can use this platform to study the single-cell genomics of a variety of diseases—potentially improving the precision and effectiveness of genetic-based diagnoses in the clinic.
FINDING MISSING LINKS
Advances in genetics and sequencing technologies have revealed many gene alterations
that are associated with disease. Despite these insights, determining causality between genetics and disease can be tedious and difficult.
“We can already sequence a child’s genome and link a DNA sequence difference to the disease,” says Grimes. “However, determining which of these DNA changes is a real disease-causing mutation is difficult, but critical to understanding the molecular mechanisms of a disease, and driving toward a cure.”
Why are these connections so tricky to identify? The more we learn about gene and protein expression in each cell at a given moment, the more complex the task becomes. Single-cell analyses are vastly increasing the number of cellular populations to be studied, and each can be affected by mutations in unique ways.
To get a complete picture of how cells work, accurate models of genetic disease are essential. By analyzing these models at a single-cell level, researchers can evaluate how mutations and medications impact different cell states.
MAPPING GENETIC CHANGES
The research team focused on identifying a gene mutation that causes severe congenital
neutropenia (SCN) in children. SCN is a blood disorder characterized by a deficiency of neutrophils, a type of white blood cell, which leads to recurrent infections and high risk of
disease.
First, researchers sequenced the genomes of cells from 225 children with SCN. They identified DNA sequence alterations in the Growth Factor Independent-1 (GFI1) gene—some known to cause neutropenia, and others of unknown clinical significance.
Next, the team created genetic models of SCN using human cells and mice to explore whether any of the new DNA changes cause neutropenia. By genetically introducing the SCN patients’ GFI1 mutations into the mouse genome, or to human induced pluripotent stem cells that generate neutrophils, they were able to study the effects in real time.
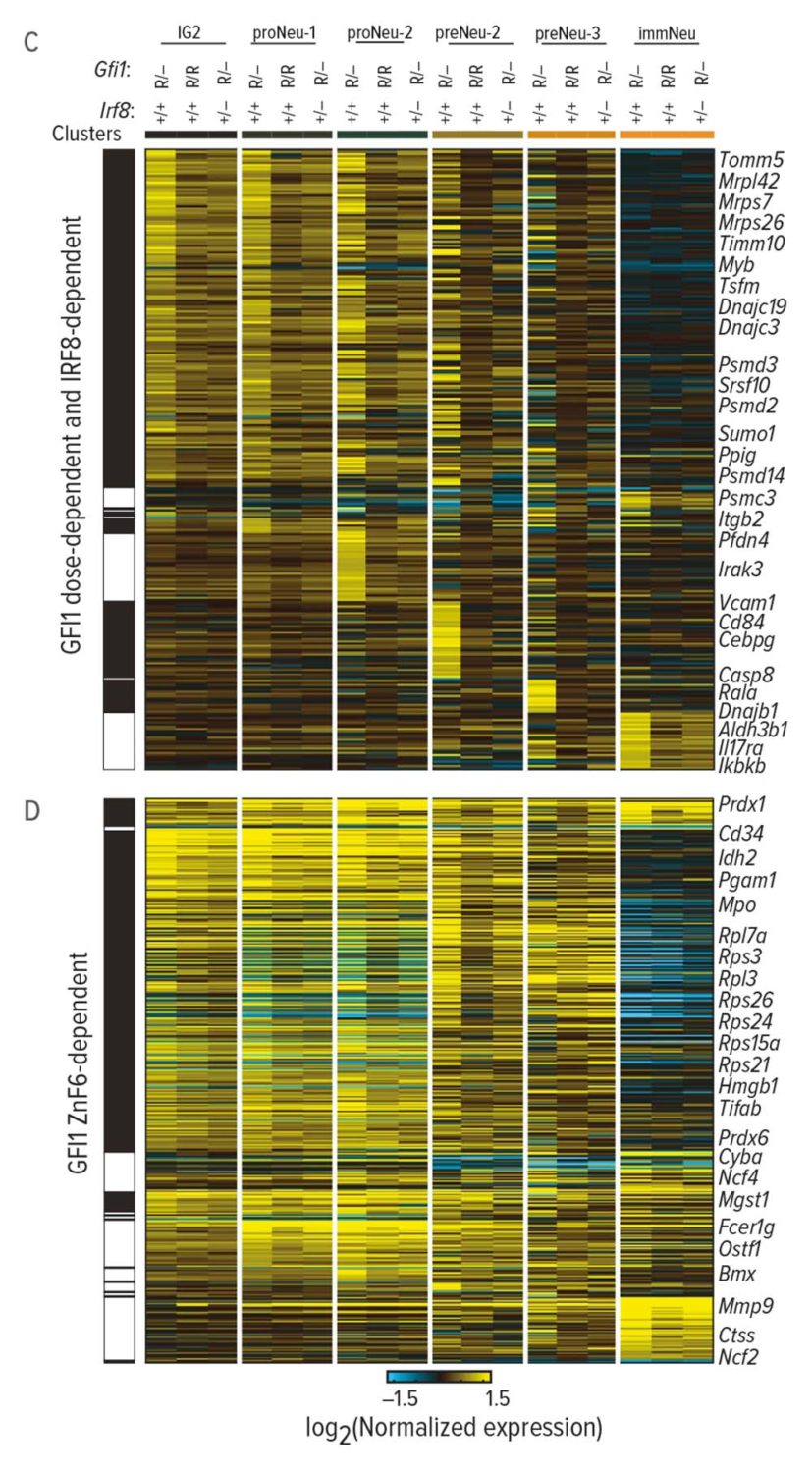
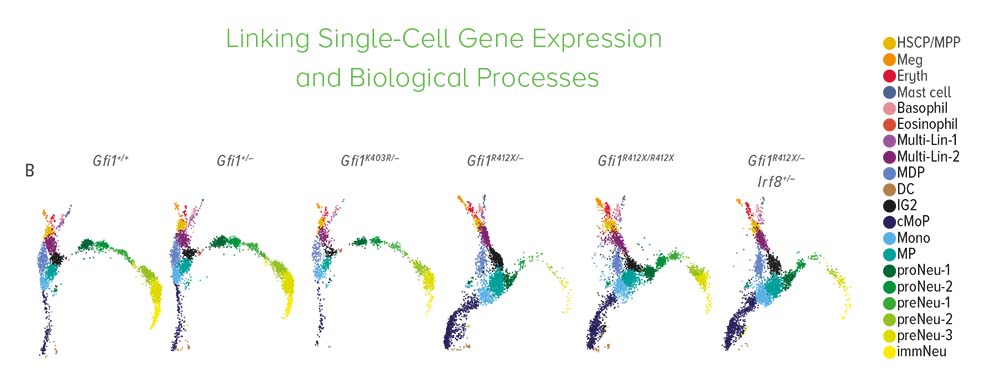
As the neutrophils developed, researchers captured the expressed genes, proteomes, and other molecular components at each stage. Using the computational tool cellHarmony, they compared downstream target genes and molecular activities of normal and mutant cells throughout neutrophil development.
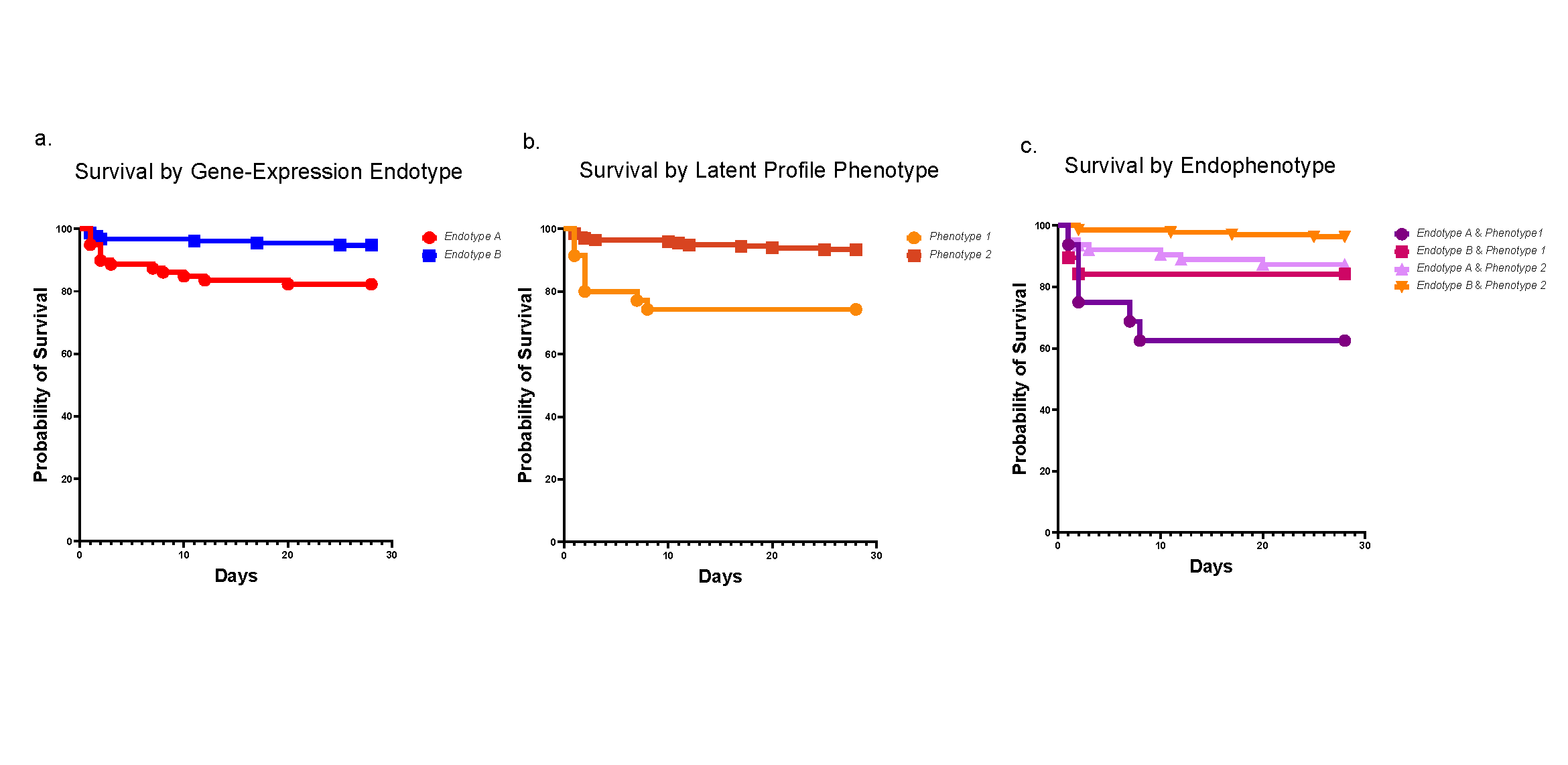
The results confirmed the disease-causing effect of GFI1 mutations. In human cells and mice, these alterations triggered molecular dysfunction that blocked the development of neutrophils.
Researchers discovered that the major problem involved chromatin in the mutant cells’ nuclei that remained open long beyond normal. This helped the team identify which cell states are most affected by the mutant gene.
“Our aim is to create a general workflow that can be applied to many diseases,” says Salomonis. “To achieve this, we leverage new genomics approaches and computational strategies to find out which mutations, cell-types, developmental periods, and molecular pathways are likely driving the disease.”
EXPLORING NEW TREATMENTS
With a better understanding of the GFI1 mutation’s impact at each stage of cell development, researchers were able to successfully treat and partially rescue neutrophils in the SCN models.
Genetic manipulations rescued the impact of the GFI1 mutations on neutrophil specification, an initial step of differentiation. But the resulting cells were still defective in commitment, a later stage of differentiation where effector functions are programmed, leaving them unable to do their job properly in the immune system. These findings emphasize the importance of evaluating the impact of mutations and therapies within each relevant cell state. Using the study’s new molecular workflow system, therapeutic interventions can be strategically targeted.
WHAT’S NEXT?
Future work will focus on understanding the steps of neutrophil commitment—specifically, how these cells program chromatin and gene expression to fight bacterial and fungal infections. Could the new single-cell and computational methods unlock insights for other diseases? With several projects in the works, researchers aim to find out.
“We are currently working on extending these bioinformatics approaches to the direct analysis of large patient cohorts,” says Salomonis. “These include funded studies of adult and pediatric hematological malignancies, neurofibroma, prenatal inflammation, ulcerative colitis, kidney transplant rejection, lung, and autoinflammatory disease.”

More Top 5 Achievements from the FY20 Research Annual Report:
-
Dose Escalation Sharply Improves Hydroxyurea Benefit for Children with Sickle Cell Anemia
-
World’s First Three-Organoid System Opens Doors for Medical Research and Diagnosis
-
Cardiac Stem Cell Therapy Improves Scar Formation Rather Than Prompting Cardiomyocyte Regeneration
-
Clinical Trial Success Caps Long Journey for HLH Treatment
Read the full report
Request a PDF
| Original title: | Mouse models of neutropenia reveal progenitor-stage-specific defects |
| Published in: | Nature |
| Publish date: | April 22, 2020 |