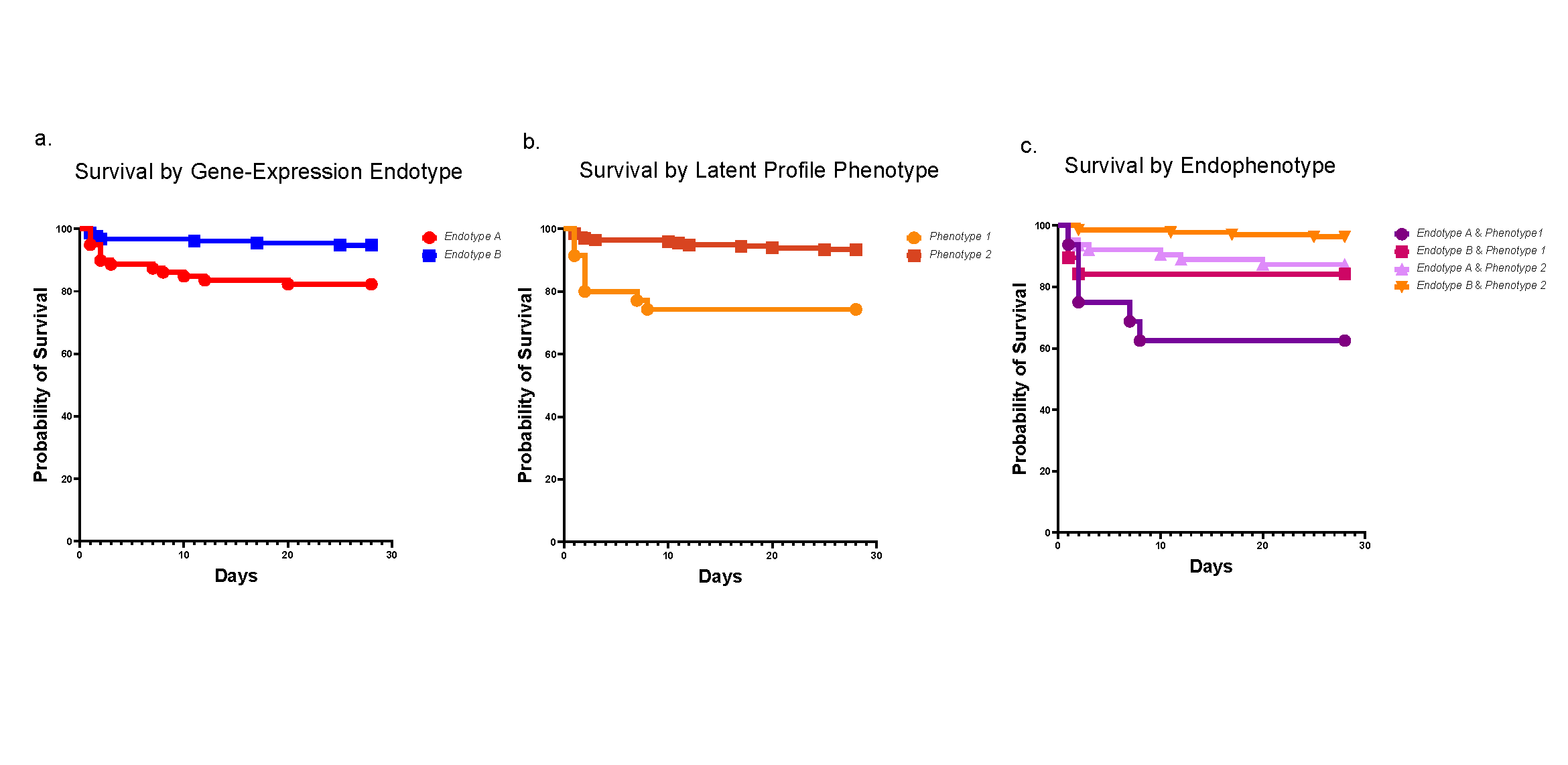
Two Genes Associated with Familial EoE
Research By: Tetsuo Shoda, MD, PhD | Marc Rothenberg, MD, PhD
Post Date: November 23, 2021 | Publish Date: Nov. 23, 2021
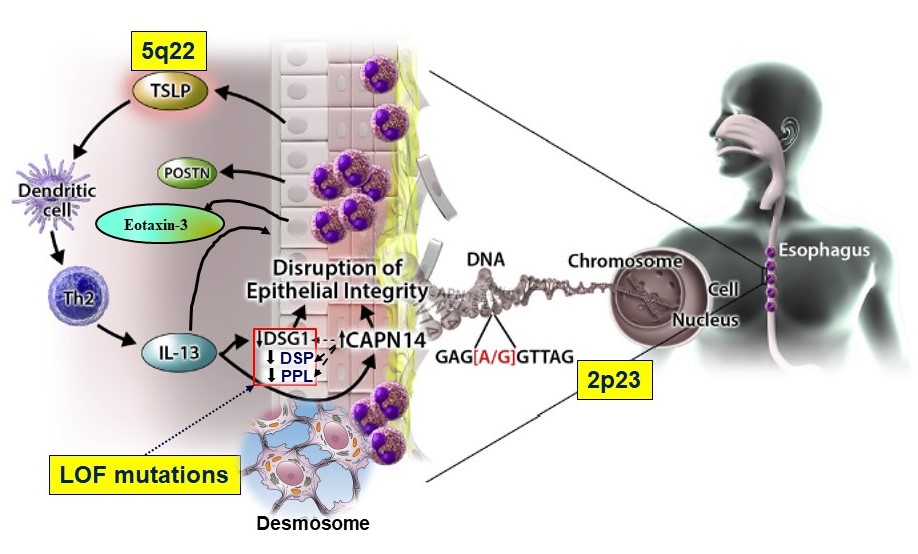
Rare variants of desmoplakin (DSP) and periplakin (PPL) appear to impair the epithelial barrier and may be acquired mutations in non-familial EoE
Experts at Cincinnati Children’s have identified two rare gene variants associated with inherited forms of eosinophilic esophagitis (EoE) that may also play roles as acquired mutations among the larger population of people with non-familial EoE.
Details about the roles played by variants of the genes desmoplakin (DSP) and periplakin (PPL) were published Nov. 23, 2021 in Nature Communications. The study was led by first author Tetsuo Shoda, MD, PhD, and senior author Marc Rothenberg, MD, PhD, Director, Division of Allergy and Immunology at Cincinnati Children’s.
The proteins generated by these genes are found in the epithelial layer of the esophagus amid structures called desmosomes that help bind cells together. These variants of DSP and PPL appear to weaken the epithelial barrier, making the tissue more prone to damage from inflammation-causing eosinophils.
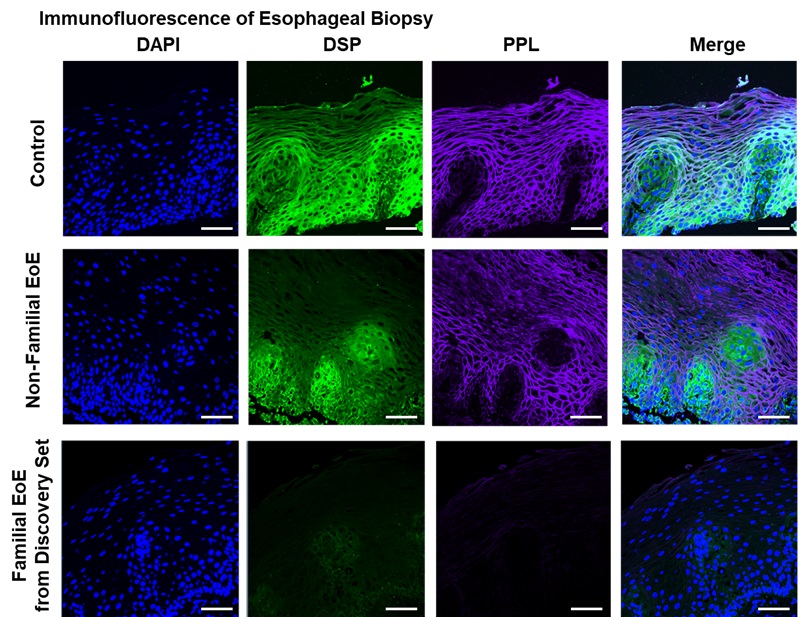
Using whole-genome sequencing, the researchers discovered the variants among five members of a family that had multiple generations of members with EoE. The team tested another 61 families with familial EoE and found 13 having either the DSP or PPL variants.
When studying rare diseases like EoE, it can be difficult to achieve the statistical power needed to identify rare gene variants that play important roles in the disease. This project overcame that challenge by starting with a set of patients that were known to be related. Now the team estimates that these gene variants account for about 21% of patients with familial EoE.
The team also suggests that finding these gene variants may lead to a new avenue of research for potential treatment.
Can EoE be treated by improving epithelial barrier function?
“Besides the impact of activated eosinophils as a functional endpoint, a growing body of evidence supports a role for altered barrier function in EoE. Dysregulation of intercellular junctions suggests a mechanism whereby inflamed and hyperproliferative esophageal epithelial tissue can be penetrated by allergens, thereby amplifying allergic inflammation,” the co-authors state. “Importantly, we present evidence that loss of DSP and PPL occurs in non-familial EoE, providing a mechanism that extends the identified pathway beyond rare familial EoE cases.”
These findings suggest that the variants could be used as diagnostic biomarkers. Also, if therapeutics targeting the desmosome can help normalize epithelial barrier function, such treatments could reduce the harmful outcomes of EoE.
About this study
This work was supported in part by grants from the National Institutes of Health (R37 AI045898, R01 AI124355, U19 AI070235, K99/R00 AI158660, and P30 DK078392); the Campaign Urging Research for Eosinophilic Disease (CURED); and the Sunshine Charitable Foundation.
Related stories:
Shoda named 2021 STAT wunderkind
EoE Research Selected as ‘Editor’s Choice’ in JACI
Cincinnati Children’s Joins NORD Rare Disease Centers of Excellence Network
$28M Grant Funds Cincinnati Children’s as Coordinating Center for Rare Diseases Research Network
| Original title: | Desmoplakin and periplakin genetically and functionally contribute to eosinophilic esophagitis |
| Published in: | Nature Communications |
| Publish date: | Nov. 23, 2021 |
Research By