Researchers Recreate Periodic Structure of Spine Development Without Biological Clocks
Research By: Ertugrul Ozbudak, PhD
Post Date: December 14, 2022 | Publish Date: Dec. 14, 2022
Developmental Biology | Top Scientific Achievement

Discovery seen as important for computational biology and bioengineering research involving disruptions to early fetal development
One of the most dramatic phases of fetal development occurs as previously unstructured collections of rapidly dividing precursor cells begin forming the embryo’s spine.
When this process goes well, it sets a solid foundation for the many other development steps that follow. When it does not, the growth defects that follow can be severe.
Now, a research team from Cincinnati Children’s and the University of Cincinnati has discovered how a set of segmentation clock genes instruct the tempo of spine formation. Mutations of clock genes lead to birth defects in humans called congenital scoliosis. The team’s findings open doors wider to a new wave of basic science research that may someday allow interventions when clock genes throughout our bodies fall out-of-sync.
Details were published online Dec. 14, 2022, in Nature.
Repairing a broken clock
In animals with spines (including humans), the growing embryo forms soft segments called somites that later form into bony vertebrae. These somites also give rise to rib formation and related back muscles and skin.
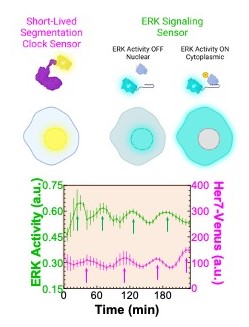
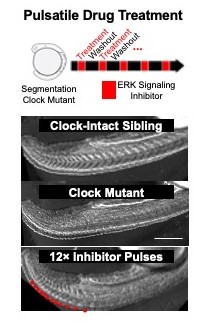
A research team led by first author M. Fethullah Simsek, PhD, and senior author Ertuğrul Özbudak, PhD, both from the Division of Developmental Biology at Cincinnati Children’s, identified a dip in molecular cell signaling that prompts the formation of a new segment (Figure 1). The work involved using zebrafish with gene modifications to detect key signaling variations. By using what they learned about the signaling, the research team was able to biochemically induce segment formation in zebrafish at will even though the fish had been engineered to lack the clock genes that normally control this process. (Figure 2).
The latest work builds on widely shared findings from the team about how co-expressed gene pairs help drive the timing of body segmentation. That study was published Dec. 23, 2020, in Nature.
“We think this study will be important for researchers in biology, bioengineering, and computational biology,” Özbudak says. “Understanding how cells are primed to form a segment boundary
at a specific location could help researchers understand the origins of other malformations that can occur during fetal development in addition to early spine formation.”
The team confirmed that the signaling molecules they studied are conserved from fish to humans. However, it will require much more research to determine if the interventions that helped correct spine malformations in the zebrafish can be translated to humans.
One hopeful longer-term application of this study may be that it provides guidance for attempting to grow segmented tissues (like the spine and digits in the hand) in the lab, suggesting a new front for organoid development.
“Broad animal species from centipedes and beetles to humans segment their body sequentially. While the molecules involved diverge dramatically among species, our study indicated that sequential segmentation can still be achieved as long as a clock stamps its periodicity on a morphogen gradient,”
Özbudak says. “We anticipate our findings will inspire engineering repetitively organized tissues in dish by utilizing pulsatile perturbation of signaling gradients.”
Next steps
The next step is to discover the molecular link between the segmentation clock and its downstream signaling pathway.
“We are hopeful that discovering the so-far missing molecular link could be clinically relevant and targetable,” Özbudak says.
About the study
This work was funded by a grant from the National Institutes of Health (R01HD103623). Other co-authors from Cincinnati Children’s were Didar Saparov, PhD candidate, and Oriana Zinani, PhD. Co-authors from the University of Cincinnati were Angad Singh Chandel, MS, and Nicholas Clason.
More 2023 Research Highlights
Chosen by the Division of Developmental Biology
Gu X, Heinrich A, Li SY, DeFalco T. Testicular macrophages are recruited during a narrow fetal time window and promote organ-specific developmental functions. Nat Commun. 2023;14(1):1439. Published 2023 Mar 15. doi:10.1038/s41467-023-37199-0
Mukherjee S, Luedeke DM, McCoy L, Iwafuchi M, Zorn AM. SOX transcription factors direct TCF-independent WNT/β-catenin responsive transcription to govern cell fate in human pluripotent stem cells. Cell Rep. 2022;40(8):111247. doi:10.1016/j.celrep.2022.111247
Pitstick AL, Poling HM, Sundaram N, et al. Aggregation of cryopreserved mid-hindgut endoderm for more reliable and reproducible hPSC-derived small intestinal organoid generation. Stem Cell Reports. 2022;17(8):1889-1902. doi:10.1016/j.stemcr.2022.06.011. Read more on our Science Blog.
Elliott KH, Balchand SK, Bonatto Paese CL, et al. Identification of a heterogeneous and dynamic ciliome during embryonic development and cell differentiation. Development. 2023;150(8):dev201237. doi:10.1242/dev.201237
Kim YS, Yuan J, Dewar A, et al. An unanticipated discourse of HB-EGF with VANGL2 signaling during embryo implantation. Proc Natl Acad Sci U S A. 2023;120(20):e2302937120. doi:10.1073/pnas.2302937120. Read more on our Science Blog.
View more discoveries from 50 research divisions and areas
Return to the 2023 Research Annual Report main features
| Original title: | Periodic Inhibition of ERK Activity Drives Sequential Somite Segmentation |
| Published in: | Nature |
| Publish date: | Dec. 14, 2022 |
Research By

Our overriding interest is to achieve a systems-level understanding of embryonic development and pattern formation by integrating quantitative experiments with mathematical modelling.