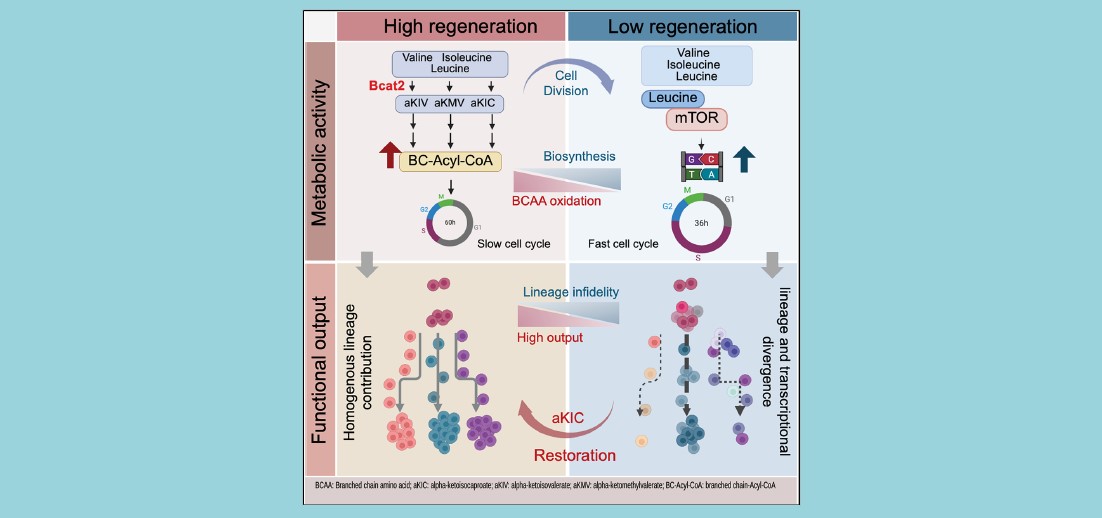
Single-Cell Web Portal Creates First Atlas and Workbench for Exploring COVID-19 Immune Host Responses
Research By: Bruce Aronow, PhD | Kang Jin
Post Date: September 27, 2021 | Publish Date: Sept. 9, 2021
“With ToppCell, we’ve created an interactive toolkit that allows researchers to see and play with gene signatures in a hierarchically organized way. Users can gain insights about immune responses of many different diseases and viruses, including COVID-19.”
—Kang Jin
When a person becomes infected with SARS-CoV-2 (COVID-19), exactly how their immune system responds can lead to outcomes encompassing life, death, and extended illness. What are the different immune responses responsible for these life-altering consequences?
Over the past nearly two years of the COVID-19 pandemic, research teams have devoted themselves to collecting samples and generating rich data from infected individuals around the world. These individuals include wide ranges of disease severity, survival, age, and clinical treatment.
Investigators have applied powerful single-cell RNA-seq technology to understand the constellations of cells present in samples of infected or secondarily affected tissues, blood, and other fluids. Using this data, investigators can learn how tissues, cells, and the immune system respond in the course of COVID-19 disease.
The power and depth of these data could help predict outcomes for many affected individuals and provide a path to improving viral immunity. However, for a variety of reasons, most biomedical researchers are unable to explore these data and realize its great potential.
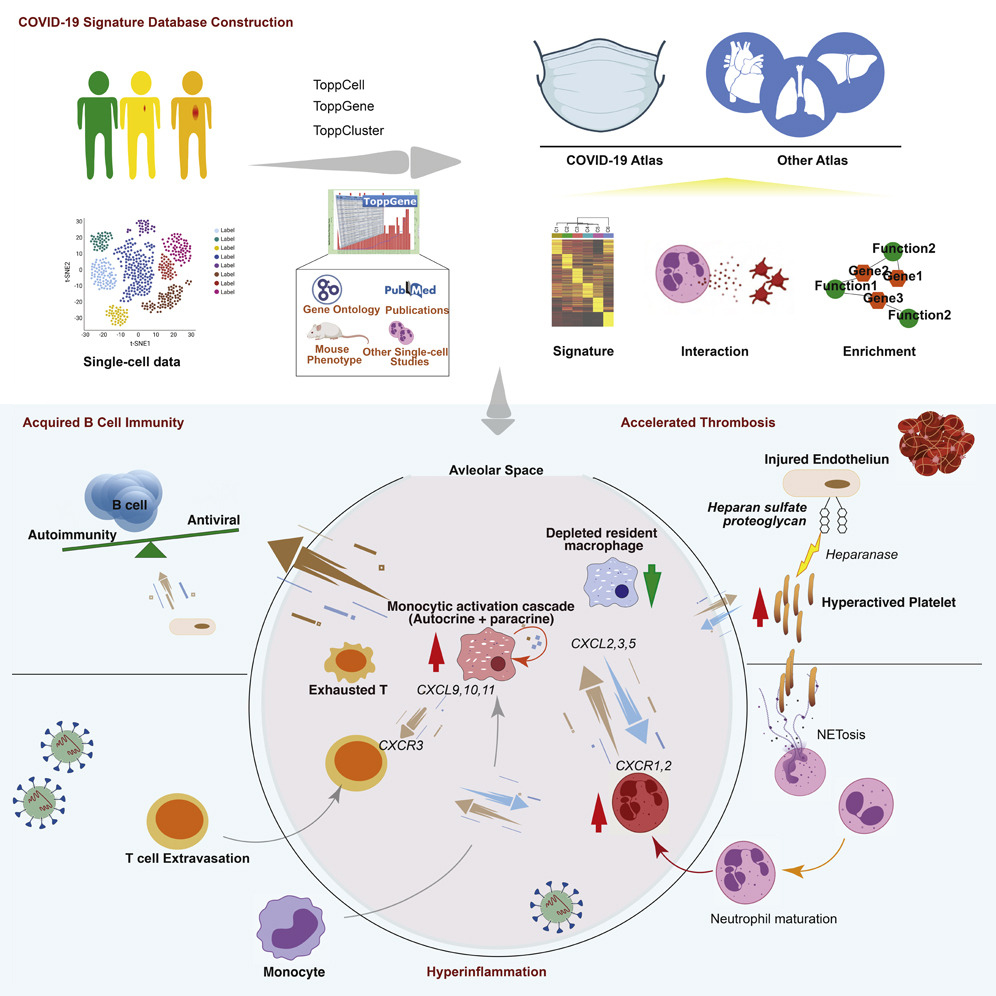
ToppCell, a new web portal designed by researchers at Cincinnati Children’s, is the first type of data mine that allows researchers to go beyond the identification of known and unknown cell types, allowing for in-depth examination of which cell types drive gene interactions and determine key signaling pathways. The tool improves how researchers can visualize and explore data to formulate hypotheses about immune responses.
Described in a study published Sept. 9, 2021, in the journal iScience, the ToppCell toolkit creates the first-ever COVID-19 immune signature atlas.
A New Way to Explore How Different Cells Function
In recent years, single-cell RNA-seq technology has transformed our ability to study biological systems. With the COVID-19 pandemic, researchers began applying this technology to learn more about how individual patients were responding to infection, generating a wealth of single-cell data.
However, interpreting and utilizing these data has been a difficult task. Beyond the conclusions made in individual manuscripts, how can we discover more about what the data is telling us?
ToppCell helps users explore data in a comprehensive and flexible way. The tool curates gene signatures of cells based on their lineages, cell classes, subclasses, and other clinical information. This generates multi-layered atlases that allow users to investigate the roles of different cell types, genes, pathways, and procedures.
“There are other tools for single-cell visualization, but these mainly focus on reduced dimensions and interaction with specific features,” says lead author Kang Jin, a PhD student in the Division of Biomedical Informatics. “For complicated data, a small list of features cannot provide a comprehensive view. With ToppCell, we’ve created an interactive toolkit that allows researchers to see and play with gene signatures in a hierarchically organized way. Users can gain insights about immune responses of many different diseases and viruses, including COVID-19.”
A Trove of COVID-19 Immune Response Data
Among COVID-19 patients, clinical outcomes are highly variable. Poorer outcomes are associated with immunological and inflammatory responses to the infection. Many recent single-cell expression profiling studies have characterized patterns of these responses, but using these data beyond markers, cell types, and individual signatures was not possible across datasets.
By offering a well-organized and systematic study of immune cells across tissues, ToppCell enables a deeper understanding of the underlying basis of COVID-19 host defense and pathobiology. In this study, researchers harmonized and analyzed eight single-cell RNA-seq datasets from COVID-19 and immunologically-related studies—in total, covering more than 480,000 human blood and lung cells.
The team used these datasets to assemble an integrated COVID-19 atlas for deriving, characterizing, and establishing reference gene expression signatures. Based on their datamining approaches, researchers proposed hypotheses of disease development that could shed light on possible treatments.
“As examples of how the datamine can be leveraged, we identified three major patterns in the data that could be related to host immune responses and mechanisms capable of driving COVID-19 severity and downstream sequelae,” says corresponding author Bruce Aronow, PhD, of the Division of Biomedical Informatics.
“In one pattern, we found striking differences among daughter cells derived from infection-activated monocytes that specifically enabled them to take on individualized roles in an amplifier network. This could both inhibit the virus directly, and recruit lymphocytes to fight the virus.
“A second pattern implicated platelet responses that had the potential to cause them to act as incendiary devices that could attack vascular endothelial cells, accelerate thrombosis, and lead to the development of thromboembolisms and pervasive vascular damage.
“A third pattern could be seen among cells exuded by the lung into bronchial airways where B lymphocytes outside of lymph nodes and the spleen were being stimulated by a platoon of cell types whose tasks seemed to converge on causing naïve B cells to generate new antibodies—on the one hand capable of attacking the virus, but on the other, because of the lack of tissue resident macrophages, strongly risk the development of post-viral autoimmunity. This is an apparently profound aspect of why so many individuals suffer from Long Haul COVID-19.”
What’s Next?
Currently, the research team has curated over 45 public single-cell studies covering various tissues in human, mouse, and in-vitro cell lines. These are categorized into eight different atlases, including the COVID-19 Atlas.
However, there are many tissues yet to be included in the database. Researchers plan to work closely with the Human Cell Atlas to build a larger curation of single-cell data, integrating other modalities including single-cell ATAC-seq and spatial data.
About the Study
Funding for this study was provided by LungMap (U24, HL148865), Digestive Health Center (P30, DK078392), Center of Excellence in Molecular Hematology (U54, DK126108), the John J Hutton Professorship in Biomedical Informatics, and the Harold C. Schott Foundation funding of the Harold C. Schott Endowed Chair, UC College of Medicine.
| Original title: | An Interactive Single Cell Web Portal Identifies Gene and Cell Networks in COVID-19 Host Responses |
| Published in: | iScience |
| Publish date: | Sept. 9, 2021 |
Research By