Researchers Produce Annotated Molecular Map of Eosinophilic Esophagitis
Research By: Leah Kottyan, PhD | Matthew Weirauch, PhD | Marc Rothenberg, MD, PhD
Post Date: January 26, 2024 | Publish Date: Dec. 27, 2023
Tool may serve as decision-making guide for biopsies and personalized treatment.
Symptoms of eosinophilic esophagitis (EoE) mirror those of many common conditions, making it challenging to diagnose. Surgical biopsy is the diagnostic gold standard, but no guidelines exist to help clinicians decide whether to perform one.
Researchers at Cincinnati Children’s hope to improve the way genetic risk can be incorporated into decision making with help from a novel, DNA-based tool called massively parallel reporter assays (MPRA). They recently published a study in the American Journal of Human Genetics demonstrating MRPA’s potential to guide diagnostic decision-making and develop personalized therapeutics.
Exploring EoE’s Genetic Component
EoE is an inflammatory allergic disease in which white blood cells called eosinophils enter the esophagus. Two decades of research from the Cincinnati Center for Eosinophilic Disorders at Cincinnati Children’s demonstrate that genetic factors play a role in determining who develops this disease.
“Cincinnati Children’s researchers have published numerous studies identifying genetic risk loci across the genome,” says Leah Kottyan, PhD, who co-authored the MPRA study with Matthew Weirauch, PhD. “Previously, we created a map showing where these genetic risk loci are found on DNA strands that include polymorphic genetic variants. Most of these variants are in non-coding spaces that do not impact the protein amino acid composition of gene products. Instead, these risk loci change transcriptional regulation—how, when and how much gene is made.”
The study was a collaboration with Marc Rothenberg, MD, PhD, director of the Cincinnati Center for Eosinophilic Disorders and the Division of Allergy and Immunology at Cincinnati Children’s, who has worked with Kottyan and Weirauch for years. The Cincinnati center includes several laboratories and patient advocacy groups that work collectively to understand the origins of EoE as well as diagnostic and therapeutic opportunities.
Annotating the Genome Risk Loci Map
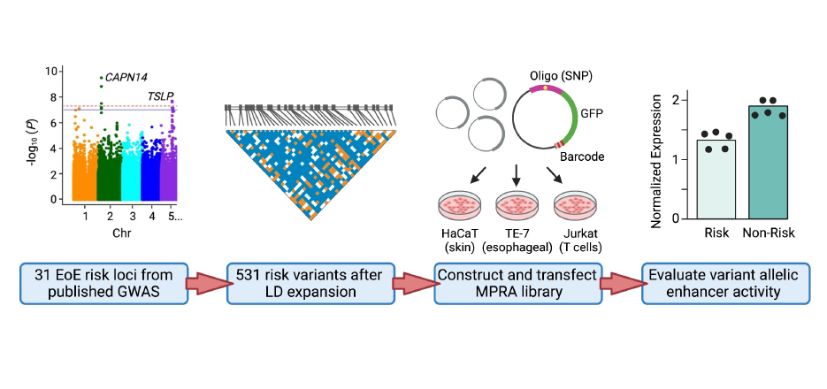
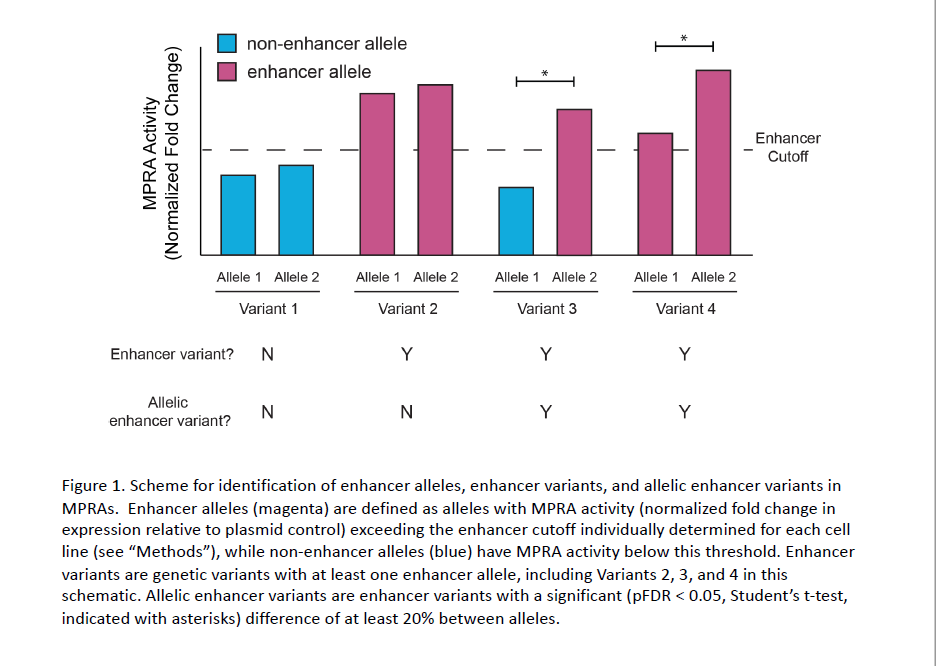
Kottyan and her colleagues began developing the MPRA molecular tool several years ago. The process starts with a droplet and uses DNA “barcodes” to measure how genetic variants change gene expression. The tool provides a comprehensive biochemical assessment of each identified EoE risk variant—essentially, it annotates the genetic risk loci map.
Using the MPRA tool, the team identified 32 EoE risk variants that change gene expression levels. By applying the MPRA to three cell types (esophageal, skin and T cell), the team identified shared- and tissue-specific ways EoE risk variants can impact gene expression.
“We prioritized the 32 genetic variants based on molecular data, out of the hundreds of genetic variants that are statistically indistinguishable from genome-wide association studies based on linkage disequilibrium,” says Kottyan, a researcher in the Division of Allergy and Immunology and the Center for Autoimmune Genomics and Etiology at Cincinnati Children’s. “Achieving this level of specificity and accuracy demonstrates the utility of using MPRA to understand genetics-dependent biology that increases a person’s disease risk.”
Study Backed by Extensive Resources
After identifying the function of the 32 EoE risk variants with genotype-dependent enhancer activity, Kottyan and Weirauch went on to identify transcription factors that mediate the genetic risk of EoE. These transcription factors include GATA3, a key regulator of allergic inflammation, and USF1, which is overexpressed in biopsies of patients with EoE.
Now validated, these transcription factors and their gene targets may become candidates for future diagnostic and therapeutic tools.
Next step: Personalized Medicine Strategies
The research team is integrating what they know about the genetic, demographic, clinical and molecular risks associated with EoE into a comprehensive model that can help physicians better diagnose and treat the disease.
Rothenberg, a renowned EoE expert, credits a culture of collaboration at the Cincinnati Center for Eosinophilic Disorders between clinicians, bench researchers, computational scientists and other experts for making studies like this one possible.
“The center brings together diverse healthcare providers and researchers who share a common goal of better understanding and treating eosinophilic diseases,” Rothenberg says. “Clinicians develop ideas that our scientists then test in the laboratory. Likewise, bench scientists partner with clinicians to test their own ideas in the patient care setting. These synergistic relationships help make Cincinnati Children’s the leader in research and clinical care innovation, as reflected in our important paper led by Dr. Kottyan and her colleagues.”
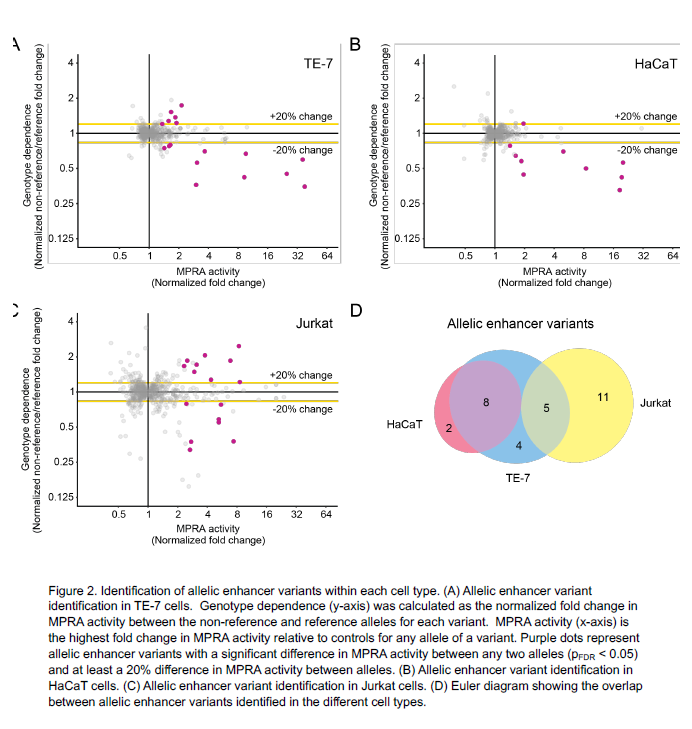
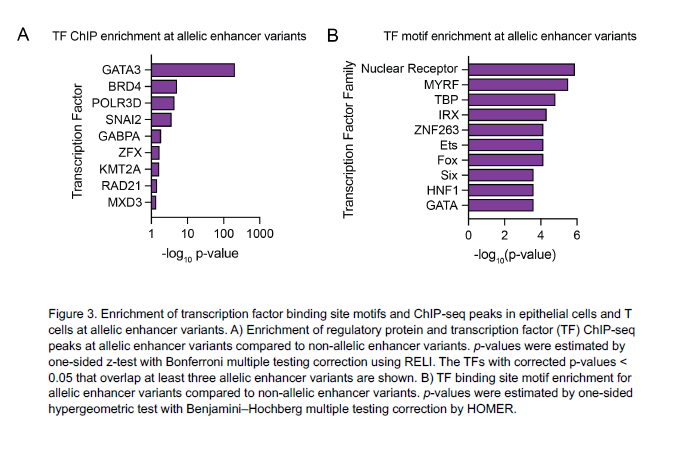
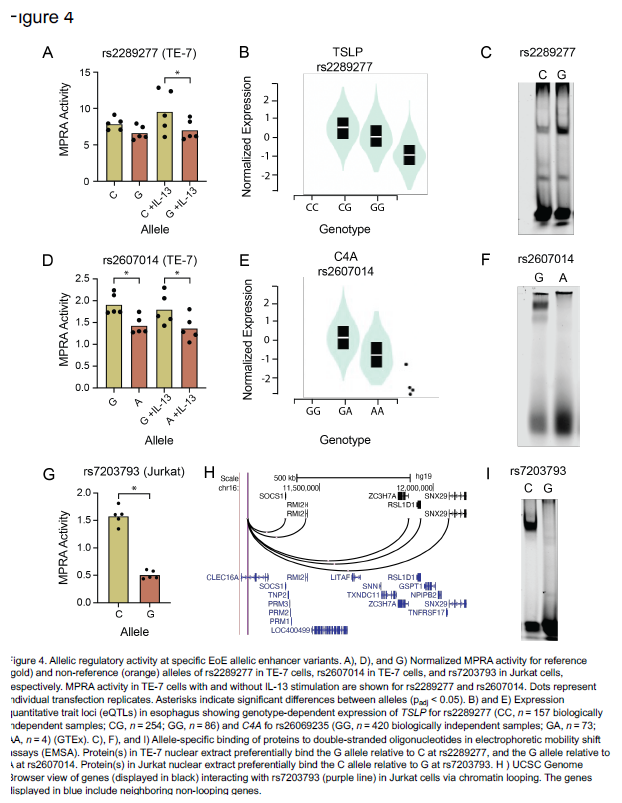
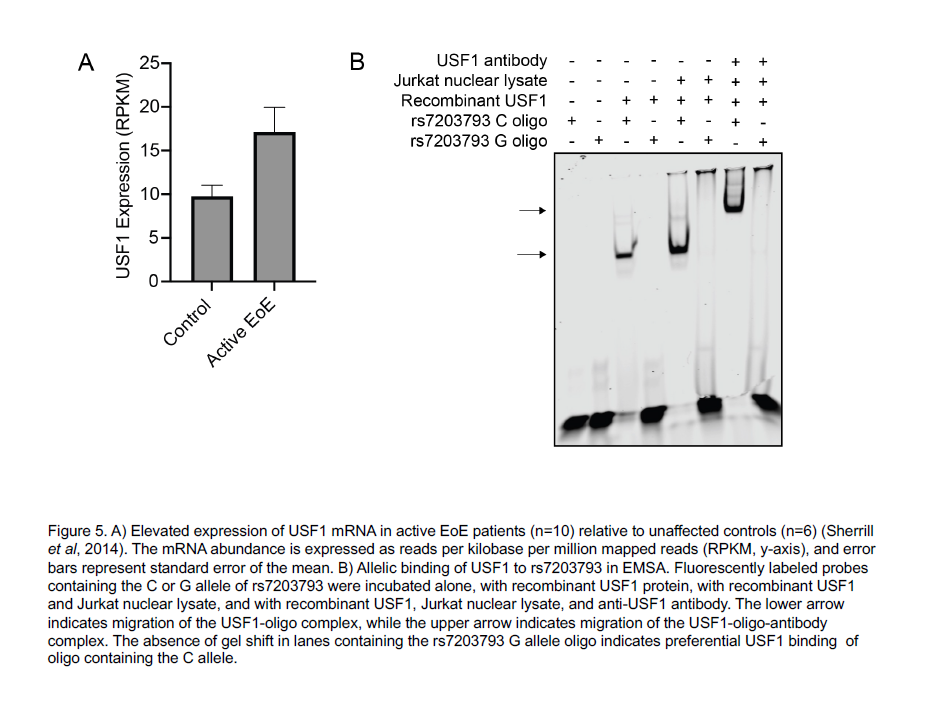
| Original title: | Systematic identification of genotype-dependent enhancer variants in eosinophilic esophagitis |
| Published in: | American Journal of Human Genetics |
| Publish date: | Dec. 27, 2023 |
Research By

My laboratory studies the genetic etiology of diseases that have an immunological component.

I am a bioinformatician geneticist who works to achieve a thorough understanding of human and viral transcriptional regulation mechanisms in complex diseases.

The Rothenberg CURED Research Laboratory, supported by the Campaign Urging Research for Eosinophilic Diseases (CURED), is focused on elucidating the mechanisms of allergic responses, especially in mucosal tissues such as the gastrointestinal tract and lung.