Nr2f Nuclear Receptors Play Key Roles in Cranial Development
Research By: Ebuka Okeke, MS | Lindsey Barske, PhD
Post Date: December 5, 2022 | Publish Date: Dec. 5, 2022
Human Genetics | Top Scientific Achievement

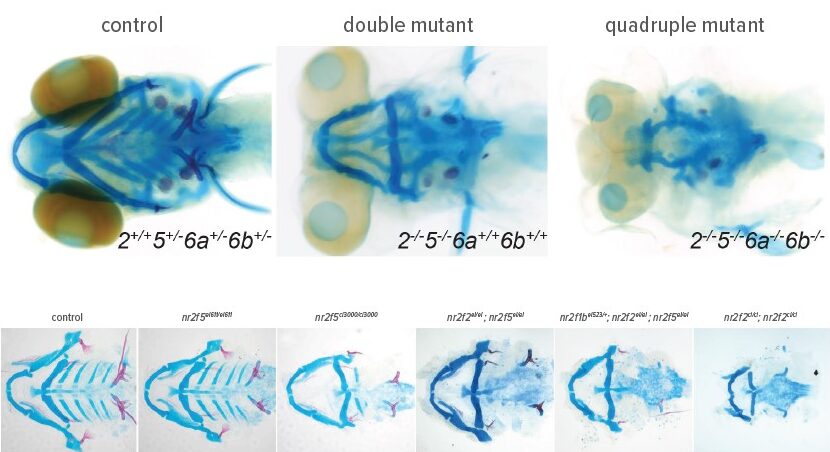
In a deep study of zebrafish craniofacial development, a team of Cincinnati Children’s researchers has identified a key activator needed to trigger a crucial transition of cranial neural crest cells (NCCs) into skeletal progenitor cells. The discovery sheds new light on formation of the face, jaw and skull bones and may someday lead to ways to improve outcomes when this development process goes awry.
The study, led by Chukwuebuka Okeke, MS, and corresponding author Lindsey Barske, PhD, zeroes in on the early-stage skeletal development role played by Nr2f nuclear receptors.
Cranial neural crest cells migrate into transient embryonic structures called the pharyngeal arches, which form the jaw and throat skeleton. As they finish migrating, Nr2f receptors activate an “ectomesenchyme” transcriptional program in these cells that helps them transition into bona fide skeletal progenitor cells. The study details how mutations in Nr2f receptor genes prevent this program from activating, thereby disrupting skeletal formation in the face.
The human disease gene sox10 is initially expressed in all neural crest cells but is particularly essential in the subpopulation that will become pigment, neural and glial progenitors. Additional experiments in this study clarified that failure to turn off sox10 in Nr2f mutants does not account for their failure to activate the skeletal progenitor program.
Importantly, the genetic activity documented in zebrafish appears to be highly conserved in human craniofacial development. Reports from the last decade have shown that children born with variants in the NR2F1 and NR2F2 genes exhibit mild facial dysmorphism along with visual, cognitive and other abnormalities.
While the new study opens doors for improved treatment, much more work lies ahead.
“We do not yet have robust small molecules capable of specifically controlling NR2F function, but these proteins are by nature ‘druggable’ and could be manipulated in the future,” Barske says.
Cincinnati Children’s co-authors on the publication include David Paulding, Sandhya Paudel, MS, and undergraduates Alexa Riedel and Conrad Phelan. Camilla Teng, PhD, with the University of Southern California Keck School of Medicine also contributed.
More 2023 Research Highlights
Chosen by the Division of Human Genetics
Du X, Glass JE, Balow S, et al. Genetic Testing in Patients with Neurodevelopmental Disorders: Experience of 511 Patients at Cincinnati Children’s Hospital Medical Center. J Autism Dev Disord. 2022;52(11):4828-4842. doi:10.1007/s10803-021-05337-6
Qu’d D, Schmitt LM, Leston A, et al. Behavioral and neuropsychiatric challenges across the lifespan in individuals with Rubinstein-Taybi syndrome. Front Genet. 2023;14:1116919. Published 2023 Jun 21. doi:10.3389/fgene.2023.1116919
Smallwood K, Watt KEN, Ide S, et al. POLR1A variants underlie phenotypic heterogeneity in craniofacial, neural, and cardiac anomalies. Am J Hum Genet. 2023;110(5):809-825. doi:10.1016/j.ajhg.2023.03.014
Solé-Navais P, Flatley C, Steinthorsdottir V, et al. Genetic effects on the timing of parturition and links to fetal birth weight [published correction appears in Nat Genet. 2023 Jul;55(7):1250]. Nat Genet. 2023;55(4):559-567. doi:10.1038/s41588-023-01343-9
Tran C, Khadkikar S, Porollo A. Survey of Protein Sequence Embedding Models. Int J Mol Sci. 2023;24(4):3775. Published 2023 Feb 14. doi:10.3390/ijms24043775
View more discoveries from 50 research divisions and areas
Return to the 2023 Research Annual Report main features
| Original title: | Control of cranial ectomesenchyme fate by Nr2f nuclear receptors |
| Published in: | Development |
| Publish date: | Dec. 5, 2022 |
Research By









