Heart Tissue in a Dish Reveals New Links Between Neurodegeneration and Heart Disease
Research By: Nathan Salomonis, PhD
Post Date: November 3, 2021 | Publish Date: Nov. 3, 2021
Findings led by experts in Seattle, San Francisco and Cincinnati suggest that some severe cases of heart failure have root causes surprisingly similar to neurodegenerative diseases like Alzheimer′s, Huntington′s and ALS.
One of the leading reasons why children and adults need heart transplants is a condition called dilated cardiomyopathy (DCM).
Over time, sometimes quite rapidly, the heart’s thick strong muscle tissue becomes thin and weak, causing the left ventricle to swell like a balloon. This makes the heart less able to squeeze efficiently, which can lead to blood clots, irregular heartbeats, and sometimes sudden death when the malfunctioning heart simply stops beating. The origins of cardiomyopathy are diverse, including viral infections, autoimmune diseases, toxic drug exposures, and dozens of gene mutations.
Now, a multi-disciplinary team of clinicians and researchers has deciphered the function of a specific genetic mutation that causes cardiomyopathy. Their findings, published Nov. 3, 2021, in Nature Communications, were made possible by growing gene-edited human heart tissue from induced pluripotent stem cells and measuring the activity, location and binding of this mutant protein.
The team was led by co-corresponding authors Charles Murry, MD, PhD, a regenerative medicine expert at the University of Washington; Bruce Conklin, MD, a genetic engineering expert with the Gladstone Institutes in California, and Nathan Salomonis, PhD, a computational genomics expert at Cincinnati Children’s.
“We hope this study will lead to broader insights that could lead to improved heart failure therapies,” Conklin says.
Cutting-edge experiments expose more of the heart’s inner workings
Over the last several decades, the research community has made many discoveries that have led to improved medications and medical devices that can dramatically extend life by slowing down the progression of heart failure. However, we still lack proven cures.
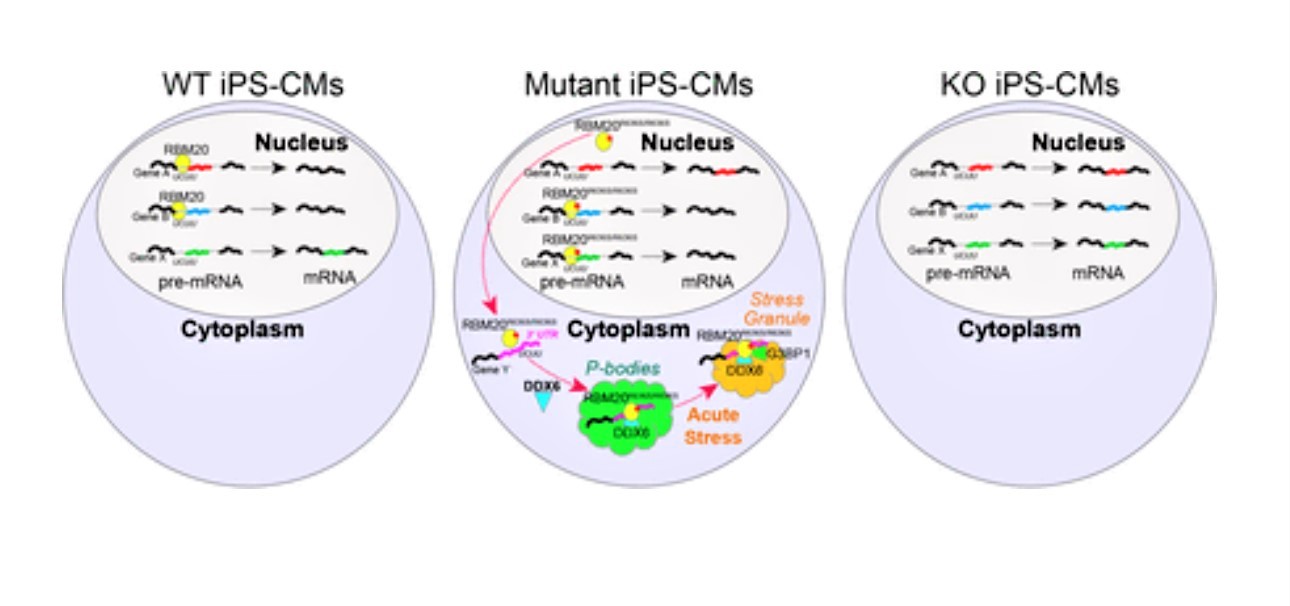
This study reveals a new mechanism of cardiomyopathy initiation by the RNA binding motif protein 20 (RBM20). This protein helps control RNA splicing in the heart, the process by which RNAs are sliced and diced to give rise to different proteins in different tissues. Normally, RBM20 splices RNAs to make proteins that enable the heart to adapt to stress and contract regularly throughout a person’s entire life. But a class of mutations in RBM20 result in severe cardiomyopathy in adulthood.
“We and others had previously studied RBM20’s function during heart development, but we had little to no clue of why it stops working in disease. We needed to step up our game if our research was to have a clinical impact,” says Alessandro Bertero, PhD, who contributed to the work while at the University of Washington and now leads an Armenise-Harvard Laboratory at the University of Turin in Italy.
Discovering this protein’s role was especially complex because knocking out this gene in animal models does not mimic the damaging effects seen in people. Instead, the work required editing the genome of healthy cells and engineering human heart tissue from these cells in a lab dish. Only by producing heart tissue similar to that found in humans could the authors understand the contractile defects and molecular mechanisms underlying this gene’s function in a controlled manner.
“That was exactly what we intended when we started this project by genome-editing induced pluripotent stem cells,” says co-leading author Yuichiro Miyaoka, PhD, of the Tokyo Metropolitan Institute of Medical Science.

First, the team observed that the engineered muscle tissue carrying the mutant form of RBM20 did not function like tissue engineered with normal RBM20 or lacking the protein all together. The mutated muscle fibers contracted with significantly less force and upstroke velocity, much like a heart affected by cardiomyopathy.
Then, at the single-cell level, the team detected another important clue. Normally, RBM20 is located exclusively within the cell nucleus. However, the mutated form localizes almost entirely out of the nucleus, in the cell’s cytoplasm.
This, by itself, did not mean much—until the cell was exposed to heavy stress. When that occurred, the mutant protein was detected within tiny “stress granules” made of protein and RNA that cells rapidly produce as a reaction to stress. In contrast, RBM20 in healthy cells remained within the nucleus and distinct from stress granules. This suggests there are additional cellular mechanisms, along with changes in splice-activity, leading to RBM20 cardiomyopathy.
“When the RNA binding landscape of mutant RBM20 was revealed by a technology called enhanced CLIP, it mimicked the binding of other splicing factors that have been implicated in neurodegenerative diseases. These factors, when mutated, also change their activity from RNA splicing to RNA aggregation outside the nucleus,” says co-author Gene Yeo, PhD, MBA, a member of the Department of Cellular and Molecular Medicine at the University of California San Diego.
“Over time, such aggregates play havoc with other cell functions, ultimately leading to the tissue-weakening of heart muscle during cardiomyopathy,” Salomonis says.
“It is intriguing to note the parallels between our observations with RBM20 and recent findings in neuro-degeneration,” the paper states. “Indeed, recent work has hypothesized cytoplasmic RBM20 may be similar to the cytoplasmic RNP granules associated with neurodegeneration (Schneider et al., 2020), such as TAU for Alzheimer′ s disease, Huntingtin for Huntington′ s disease, and FUS for amyotrophic lateral sclerosis (ALS).”
Next steps
Co-authors for this study also included scientists from the University of Cincinnati Department of Electrical Engineering and Computer Science, Sana Biotechnology, and the University of California San Francisco.
The co-authors say the 3D heart tissue model they’ve developed has the potential to be used to test new drugs to block the formation of cytoplasmic granules as a possible treatment for cardiomyopathy, even those without RBM20 mutations.
“RBM20 has been a frustrating protein to study, as animal models don’t fully recapitulate human disease pathology,” says lead author Aidan Fenix, PhD. “It’s exciting to now have an in vitro human cell model of RBM20 cardiomyopathy that shows the major clinical feature of dilated cardiomyopathy–reduced contractile force. We hope these models will speed the discovery of therapies to treat RBM20 dilated cardiomyopathy.”
About this study
This work was supported by grants from the National Heart, Lung, and Blood Institute (U01 HL099997, P01 HL089707, R01 HL130533, F32 HL156361-01, HL149734, R01 HL128362, R01 HL128368, R01 HL141570, R01 HL146868); the National Institute of Diabetes and Digestive and Kidney (U54DK107979-05S1); the National Science Foundation (NSF CMMI-1661730); a JSPS Grant-in-Aid for Young Scientists, and grants from NOVARTIS, the Mochida Memorial Foundation, SENSHIN Medical Research Foundation, Naito Foundation, Uehara Memorial Foundation, a Gladstone-CIRM Fellowship, and the A*STAR International Fellowship.
| Original title: | Gain-of-function cardiomyopathic mutations in RBM20 rewire splicing regulation and re-distribute ribonucleoprotein granules within processing bodies |
| Published in: | Nature Communications |
| Publish date: | Nov. 3, 2021 |
Research By