POLYseq Tool Sharply Improves Barcoding in Single-Cell Analysis
Research By: Andrew Dunn, PhD | Takanori Takebe, MD, PhD
Post Date: August 26, 2021 | Publish Date: Aug. 26, 2021
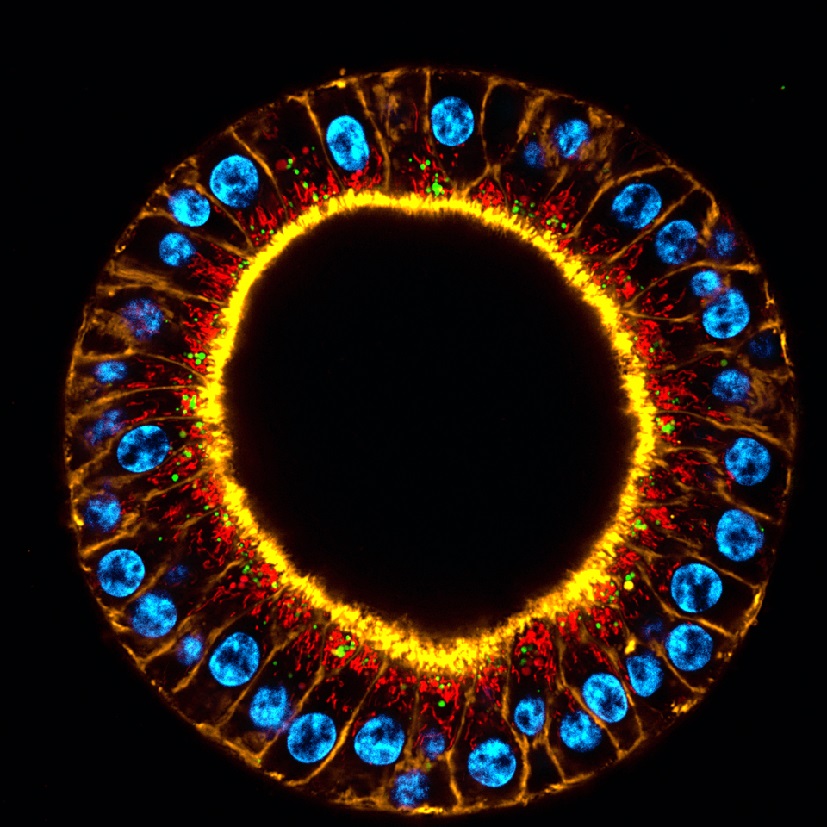
As the explosion of single-cell RNA sequencing technology continues, finding a way to track genomic activity in real time as cells morph and multiply during tissue development has emerged as a vital task for scientists seeking to understand the mechanisms of disease.
Now, a team of experts at Cincinnati Children’s has developed a tool called POLYseq that appears to make this extremely complex task much easier. Details were published Aug. 26, 2021, in the journal Stem Cell Reports. The study was led by first author was Andrew Dunn, PhD, and corresponding author Takanori Takebe, MD, PhD.
The POLYseq tool provides a set of vectors that can bind to cells of interest within five minutes. The tool is so fast that it allows scientists to observe the step-by-step transformation of cells within a liver organoid as it grows in a lab dish.
This new level of precision can support a wide range of investigative opportunities—and uses commercially available reagents, making the process cost-efficient, Takebe says.
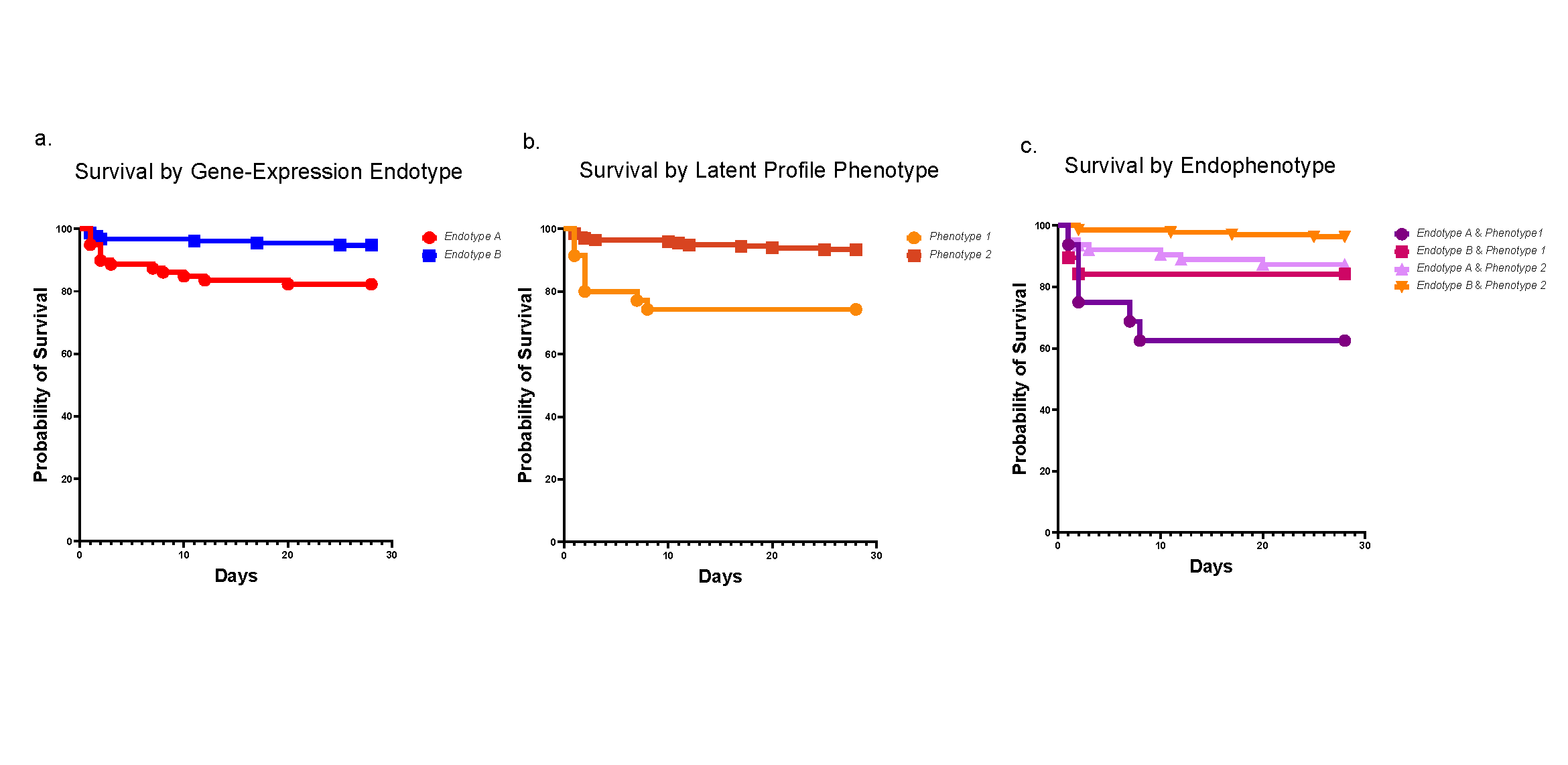
“This labeling speed provided the opportunity to rapidly barcode single-cell suspensions of diverse cell types and allowed for successful identification of directly pooled distinct human and mouse cell lines solely by barcode reads, providing a highly competitive hashing strategy compared with antibody-based methods without the restriction of specific target antigens,” Takebe and colleagues write.
POLYseq also allows for direct exclusion of doublets – overcoming a challenge for other barcoding methods – and allows investigating gene expression patterns within pooled cells that appear to have highly similar transcriptomes, the team reports.
Dunn and Takebe are listed as inventors for POLYseq-related intellectual property. Supplemental information can be found online at https://doi.org/ 10.1016/j.stemcr.2021.07.020. Researchers can contact Takebe for further information.
| Original title: | POLYseq: A poly(ß-amino ester)-based vector for multifunctional cellular barcoding |
| Published in: | Stem Cell Reports |
| Publish date: | Aug. 26, 2021 |
Research By