Molecular Signature Revealed for Placenta Hypermaturity
Research By: Heather Brockway, PhD
Post Date: November 19, 2019 | Publish Date: Nov. 8, 2019
Finding May Help Explain a Number of Previously Unexplained Preterm Births
Over the years, scientists have pinned down several factors that can trigger preterm birth, including infections, smoking and other unhealthy lifestyles, advanced age, preeclampsia, and other indications. But as many as half of preterm births are considered idiopathic (unknown cause).
Now, a study published Nov. 8, 2019, in PLOS One describes a molecular signature for a condition that may explain a number of previously unexplained preterm births: placenta hypermaturity. Here, lead author and corresponding author Heather Brockway, PhD, a Research Associate in the Division of Human Genetics, discusses the breakthrough which was co-authored by Louis Muglia, MD, PhD and Helen Jones, PhD.

What are the key take-aways from this study?
Our study provides insight into potential molecular mechanisms of idiopathic spontaneous preterm birth (isPTB). Few transcriptomic studies have been conducted on spontaneous PTB placental villous tissue.
What makes this study novel is that by conducting pairwise comparisons between isPTB, term, and gestational age controls, I was able to isolate an isPTB transcriptomic signature of about 170 candidate genes, with many being significantly upregulated compared to the gestational age controls, suggesting a hypermaturity of the placenta.
Differences in placental hypermaturity have been identified in previous histopathological studies of PTB, but this is the first time a potential molecular signature of maturity has been detected.
What did you learn about this signature?
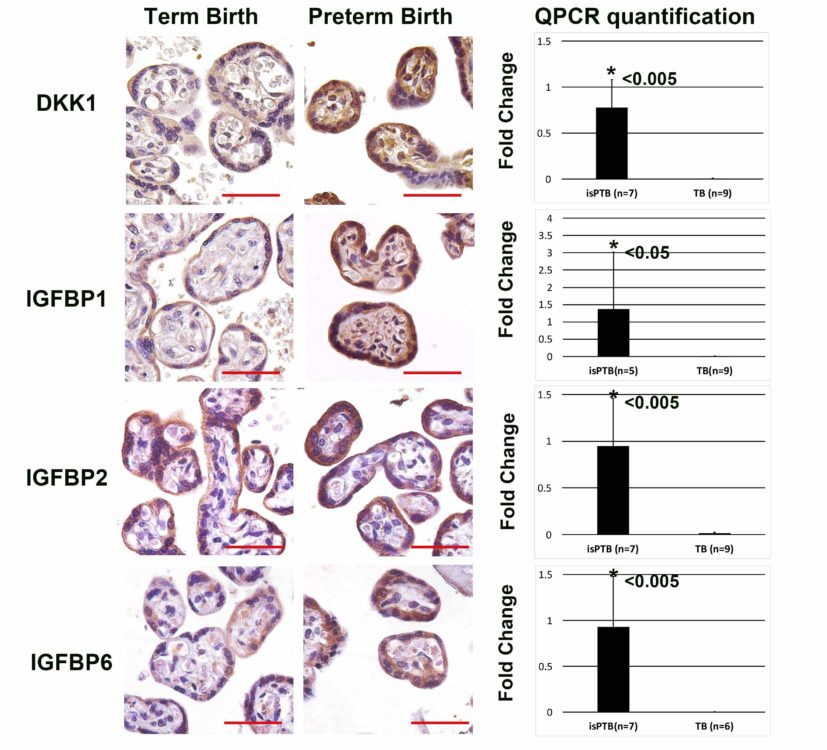
The most significant pathway identified was IGF signaling regulation, with an overrepresentation of IGFBP1, IGFBP2, IGFBP6 which can potentially turnoff IGF signaling by modulating IGF1 and IGF2 bioavailability. The IGF signaling pathway has been extensively studied in the context of fetal growth restrictions, but interestingly the fetal growth in this cohort was not affected.

This finding is particularly intriguing because in normal pregnancies, fetal and placental maturation are coupled. During the latter half of gestation, the placental maturation includes extensive remodeling of the fetal vasculature and expansion of the syncytial surface to accommodate nutrient and oxygen transfer to the maturing fetus.
These data suggest there is a developmental disconnect, and the co-maturation is now uncoupled with no effect on the fetus, only the placenta.
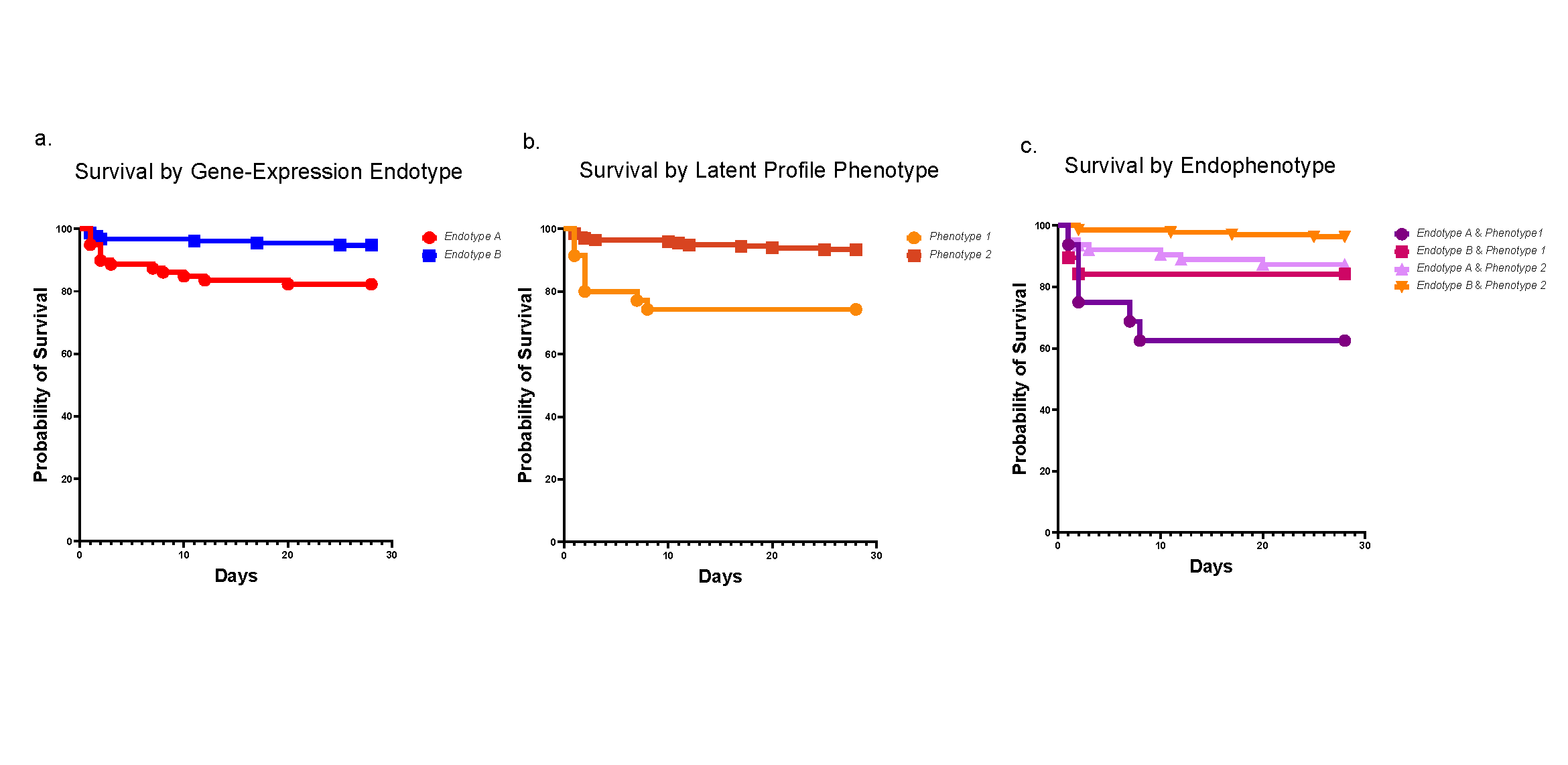
Furthermore, the gestation age controls had a unique signature of their own, one of mitotic division and cell cycling suggesting appropriate maturation for the gestational age. Remember, the placenta is supposed to be remodeling and maturing, processes that require cell cycling and division. More importantly, the isPTB placentas of the same gestational age did not share this signature, instead reflecting a signature more like that of term placentas, suggesting their maturation was finished.
Together these data suggest these isPTB placentas have aberrant maturity, in this case a hypermature phenotype leading to an earlier birth timing than normally maturing placentas.
What does this finding mean for currently expectant mothers?
This is a small preliminary study and needs to be replicated with much larger cohorts. Yet, it illustrates the power of transcriptomics to more specifically define preterm birth types at a molecular level. It is the first step in the journey of identifying the specific differences between infection associated and truly idiopathic spontaneous preterm birth.
Unfortunately, the translation of these findings into a useful tool for clinicians takes time. By creating a molecular snapshot of what is happening at delivery in term, preterm, and gestational age control placentas, we can start to unravel the molecular underpinnings of both normal and preterm birth.
What’s next with this line of research?
To do it all again, in larger, precisely defined cohorts. This cohort was quite small and that was by design to eliminate many variables that could potentially confound detection of the signatures. I’m currently applying for additional funding from the NIH Human Placental Project to expand this study into larger cohorts to validate the signatures identified and to correlate the signatures to hypermaturity morphological phenotypes. The key is to build collaborations with other preterm/placental researchers who have placental samples available for study. Few preterm cohorts exist that also have placental samples available for study.
It appears that the placenta transcriptome likely reflects its output, consisting of extracellular vesicles, cell-free nucleic acids, and proteins, all of which can be measured in maternal blood or other biofluids. The histopathological studies indicate multiple morphological phenotypes of preterm birth placentas including hypermaturity, each likely to have specific transcriptomic signatures and output.
If we can link those morphological phenotypes to transcriptomes and output then we will may be able to look longitudinally at placental maturity through what is secreted into maternal blood. This is precisely the premise behind the NIH’s Human Placental Project, which funded this initial study, along with the March of Dimes Ohio Collaborative and the Gates Foundation.
What inspired you to study preterm birth?
There are many reasons, but mostly far too many friends have had a preterm birth and they have shared their stories with me, especially the fear of losing their baby.
While neonatal medicine has come a long way and we now save so many babies, there are still parents who leave a NICU without their little one. When I came to Cincinnati Children’s, we had a motto: Change the outcome. That’s exactly what I intend to do.
—Article written by Healther Brockway, PhD, and Tim Bonfield
| Original title: | Unique transcriptomic landscapes identified in idiopathic spontaneous and infection related preterm births compared to normal term births |
| Published in: | PLOS One |
| Publish date: | Nov. 8, 2019 |
Research By