How “ceSAR” Picks the Best Molecules from a Drug Discovery Salad – Really Fast
Research By: Alexander Thorman, PhD | Andrew Herr, PhD
Post Date: August 30, 2024 | Publish Date: Aug. 30, 2024
Open access research tool created at Cincinnati Children’s and the University of Cincinnati can be up to 48,000 times faster than current methods for sifting promising “hits” from huge libraries of small molecules
One reason why successful drug development takes so long and costs so much is that it often takes lab teams a long time to sort through massive data libraries to assemble lists of seemingly promising small-molecule drug candidates only to find out that most of them cannot do the job.
Unpredicted toxicities. Off-target effects. Difficulties delivering the drug past cellular membranes. Wild variations in metabolization, and so on. Even when armed with powerful computers to help crunch the numbers, it can take years for experts in biochemistry and structural biology to whittle down hundreds of thousands of interesting possibilities to just a few that merit deeper testing.
Now, experts at Cincinnati Children’s and the University of Cincinnati College of Medicine have developed a drug discovery tool dubbed “ceSAR” that dramatically reduces the time needed to complete the sorting process while also returning better short lists of highly promising “hits.”
Details about how the new tool works—and how scientists anywhere in the world can access it—were published online Aug. 30, 2024, in the journal Science Advances.
Used one way, the tool can be 48,000 times faster than a more traditional “docking screen” approach for ranking drug candidates. Used in a more nuanced way, the tool can still be hundreds of times faster than traditional methods, but also return much stronger short lists of hopeful drug candidates.
“For example, in this study we were searching for small molecules that could block the function of a protein that prevents certain over-active immune cells from standing down,” says the study’s co-lead author co-lead author Alexander Thorman, PhD, a postdoctoral fellow with the UC Department of Environmental & Public Health Sciences. “Essentially, on this project I was running the equivalent of 75 desktop computers around the clock for about three months. But with ceSAR, those same results can be generated in less than a minute.”
Two more scientists were co-lead authors: James Reigle, PhD, a postdoctoral research fellow with the Division of Biomedical Informatics at Cincinnati Children’s, and Somchai Chutipongtanate, MD, PhD, with the UC Department of Environmental & Public Health Sciences.
Andrew Herr, PhD, Division of Immunobiology at Cincinnati Children’s, was a co-corresponding author along with Jarek Meller, PhD, a member of both the Division of Biomedical Informatics at Cincinnati Children’s and the UC Department of Environmental & Public Health Sciences.
Immediate progress in one drug candidate hunt, many more to come
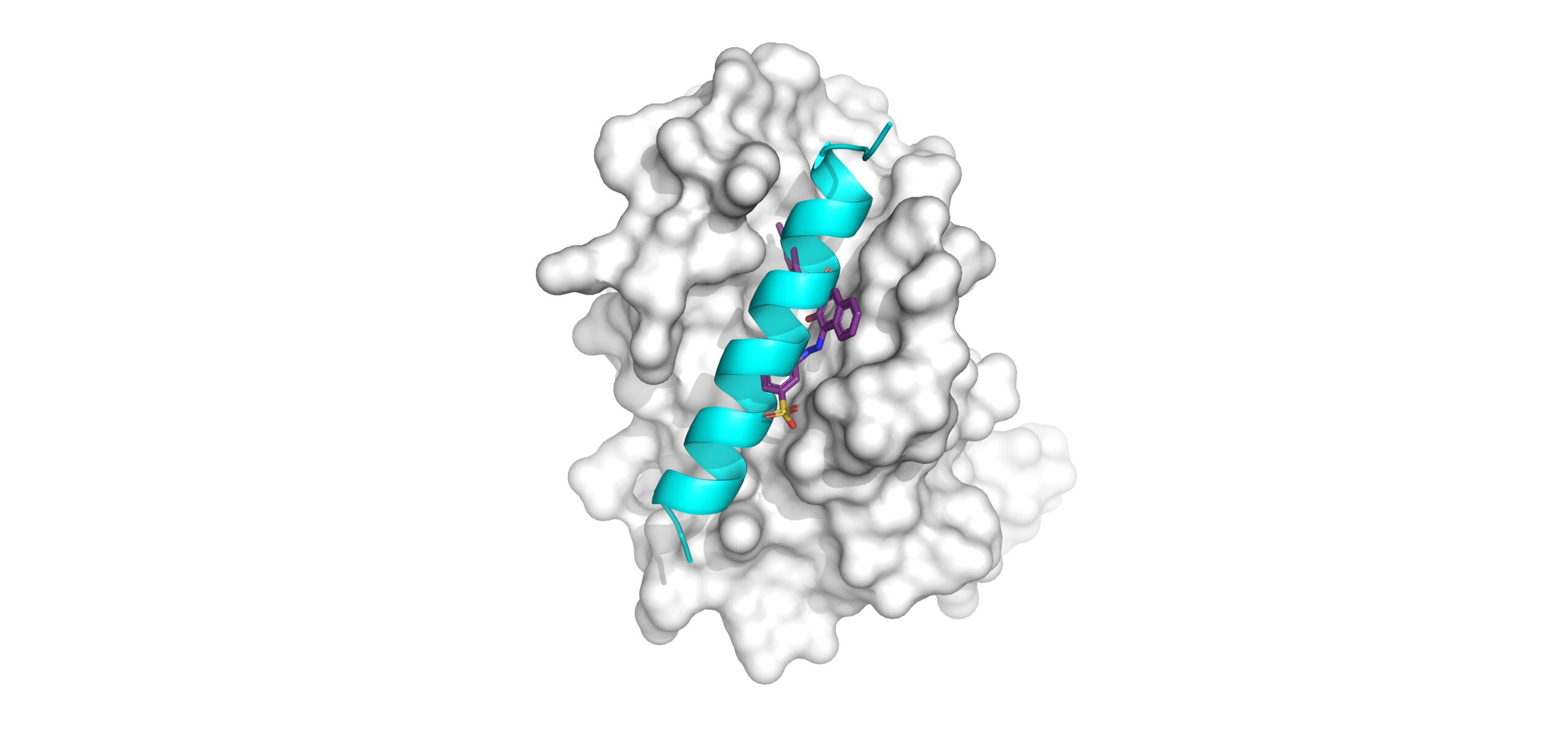
The research team demonstrated the utility of ceSAR by using it to rapidly identify small molecules that can inhibit the function of a protein called BCL2A1. Then the team used slower methods to validate that two top molecules identified in the fast process worked as predicted.
This protein is known in the science world as “an important antiapoptotic target.” It contributes to multiple diseases including treatment-resistant melanoma and preterm birth–associated inflammation.
Put another way, the BCL2A1 protein prevents T cells from dying off in a process called apoptosis when the body’s immune system no longer needs them to respond to a threat. This protein functions like a magnet that captures a tiny bullet—called a BH3 helix—that the body uses to kill the T cells.
Previous research conducted at Cincinnati Children’s by David Hildeman, PhD, and colleagues uncovered BCL2A1’s surprising power. Then the team at UC and Cincinnati Children’s used the new ceSAR tool to find a small molecule that binds to a very specific location on the BCL2A1 protein, turns off the magnet, and lets the bullets fly free to punch holes in the unwanted T cells.
“So the hope is that we can tip the balance,” Herr says. “If you have an autoimmune or an inflammatory condition that has activated immune cells going on the rampage, we could give this compound and the BH3 helix can kill off the activated immune cells—but not interfere with the other resting parts of the immune system. In that way, we hope we can calm down the activated cells without making people vulnerable to every other infection that comes along.”
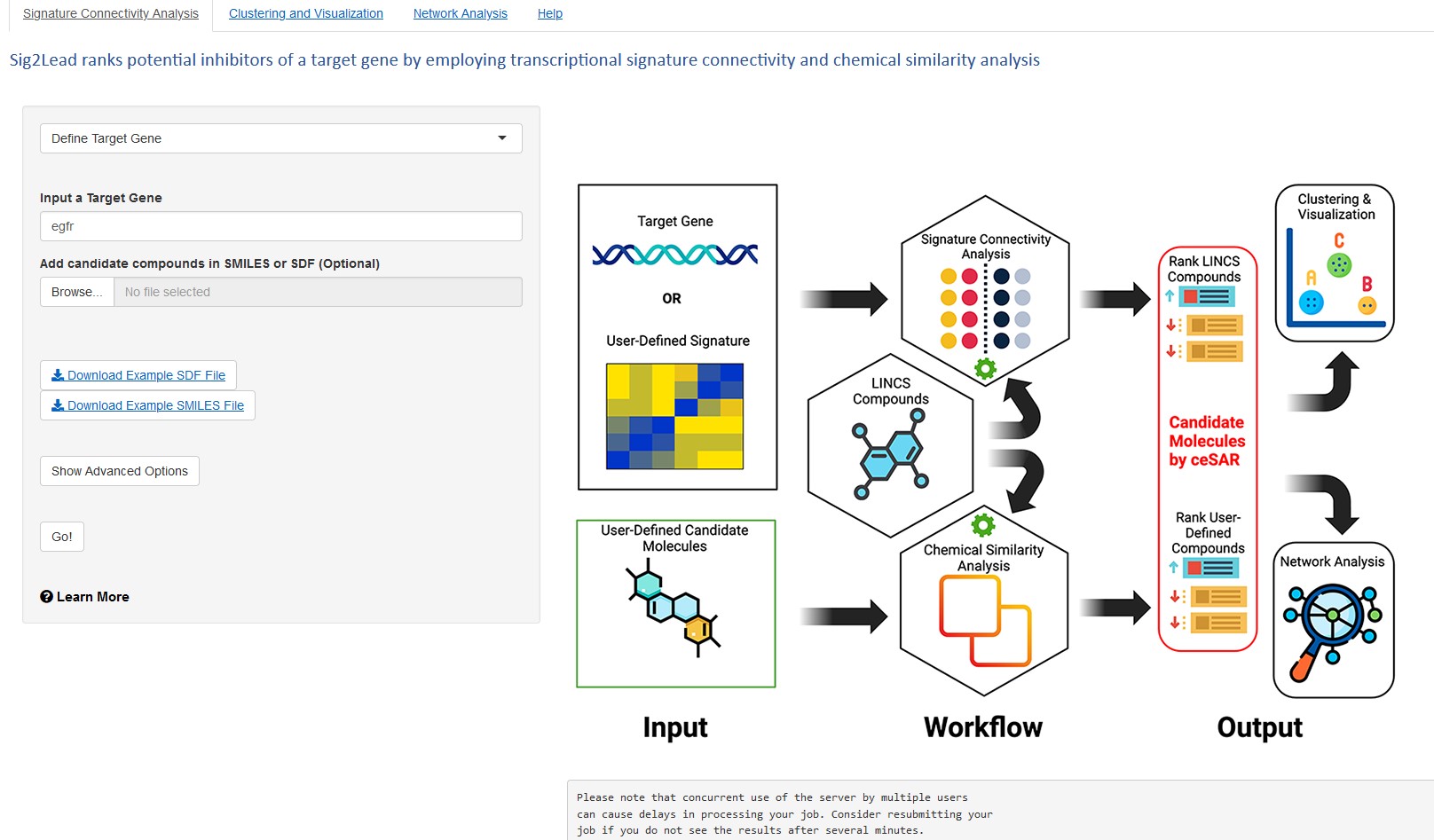
How ceSAR speeds up the search
The new ceSAR tool combines two sets of information in a novel way that shortcuts tedious searches for promising small-molecule compounds.
The process starts with the Library of Integrated Network-based Cellular Signatures (LINCS), a widely shared collection of genetic effects documented for more than 21,000 drug-like molecules. The shortcut involves comparing those genetic signatures to similar information gathered from a growing list of genes (now exceeding 4,400) that researchers have modified at various times to either gain or lose specific protein functions.
“Rather than conducting detailed laboratory tests on every compound revealed in a LINCS search, if a matching transcriptional signature emerges between these data sets, it becomes much more likely that the small molecule will induce the desired effect,” Thorman says.
Instead of tediously opening every book in the library to find all the books that might be spy novels—including boring out-of-date ones and some that might really be romance novels masked in a spy setting—the ceSAR tool almost immediately eliminates all the history books, the children’s books, and the self-help books to guide the reader to a stack of known, best-selling spy novels. This allows the reader to browse the short stack more deeply with sharply improved odds of finding a really solid spy novel.
“This is a powerful shortcut that could save enormous amounts of time and money,” Herr says. “There are many times when you get deeper into the weeds of drug discovery, you realize that this compound looks good on the bench, but it has other issues. By working with a larger pool of strong candidate compounds, you are more likely to get something that actually will make it further along the drug development pipeline.”
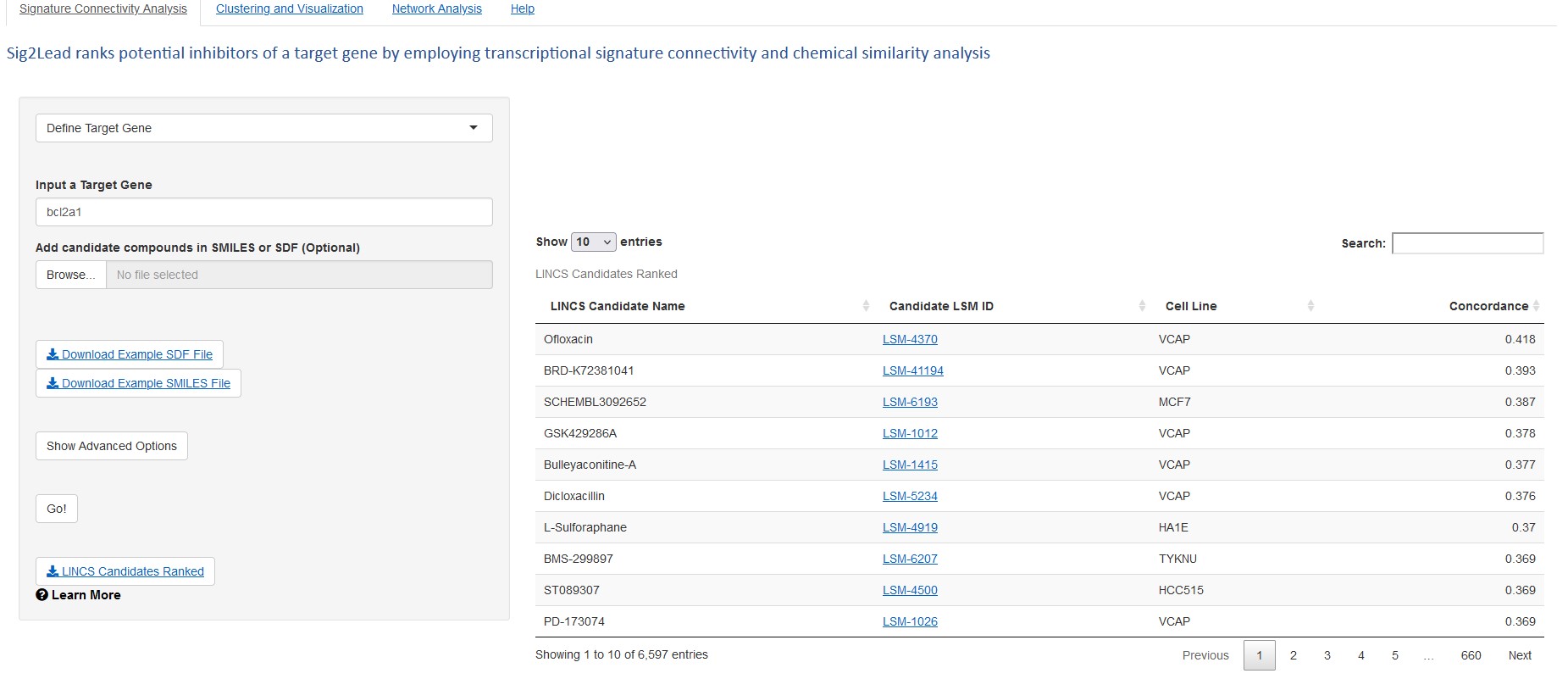
This accelerated form of virtual drug screening is important because laboratory validation testing requires many hours of work and thousands of dollars in lab supplies for each dead-end in the search.
“Instead of having to validate 100 to 1000 compounds, you only have to test between one and 10,” Thorman says.
Next steps
This research has already resulted in one US patent (#11,541,073 B2) granted Jan. 1, 2023; and two pending US patent applications (#17/597,515 and #18/083,617) both filed Dec. 19, 2022. Related drug development work continues.
Meanwhile, the ceSAR tool is publicly available at no cost to the science community to use to investigate other compounds for other conditions. An app can be downloaded from https://github.com/sig2lead. Users also can find the tool on a web server at http://sig2lead.net.
About the study
Several Cincinnati Children’s and UC experts contributed to this study and have been listed as co-authors: Juechen Yang, PhD candidate, Behrouz Shamsaei, PhD, Marcin Pilarczyk, PhD, Mehdi Fazel-Najafabadi, PhD, Michal Kouril, PhD, Surbhi Bhatnagar, PhD candidate, Sarah Hummel, MA, Wen Niu, PhD, Ardythe Morrow, PhD, Maria Czyzyk-Krzeska, MD, PhD, William Seibel, PhD, Nicolas Nassar, PhD, Yi Zheng, PhD, David Hildeman, PhD, and Mario Medvedovic, PhD.
Experts with the University of Toledo (Ohio) and Nicolaus Copernicus University (Poland) also contributed.
Funding sources for this work included grants from the National Institutes of Health (U54 HL127624, P30 ES006096, R01 MH107487, R01 CA122346, R01 GM128216, T32 CA236764, R01 CA237016, R21 HD090856, and UL1 TR001425), and awards from the Veteran’s Administration, the University of Cincinnati Cancer Center, and the Cincinnati Children’s Innovation Fund.
| Original title: | Accelerating drug discovery and repurposing by combining transcriptional signature connectivity with docking |
| Published in: | Science Advances |
| Publish date: | Aug. 30, 2024 |
Research By


Our laboratory is interested in mechanistic studies of cellular adhesion and receptor-mediated signaling in immunology and microbiology.